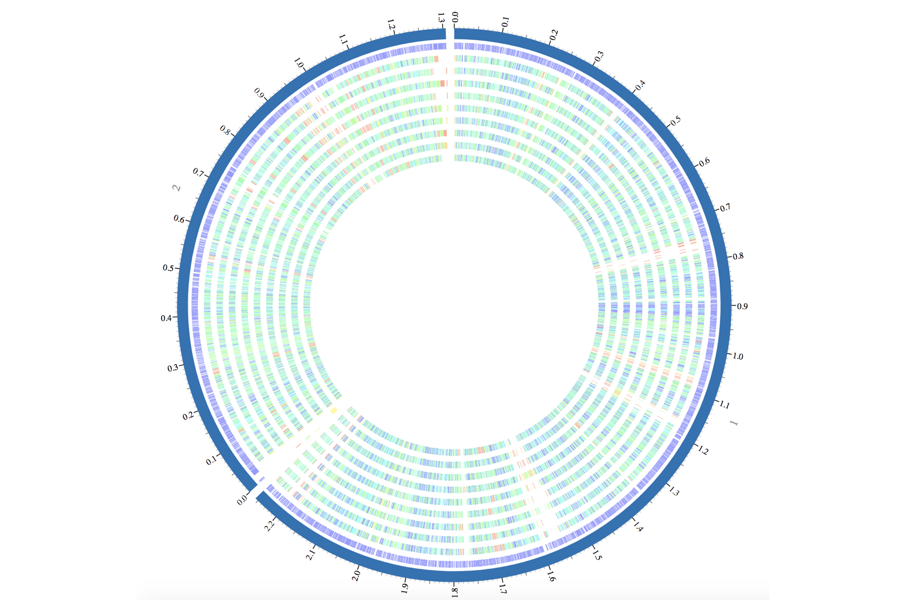
University of Chicago computer science professor Rick Stevens and J. Craig Venter Institute bioinformatics director Richard Scheuermann were awarded funding to launch the Bacterial and Viral Bioinformatics Resource Center (BV-BRC), a new online tool that merges two databases: the Pathosystems Resource Integration Center (PATRIC), a bacterial resource, and the Virus Pathogen Resource (ViPR).
This is part of a larger National Institute of Health–funded push towards greater consolidation of bioinformatic resources. While the bioinformatics boom has provided a wealth of information, its abundance has also become overwhelming. BV-BRC is part of a consolidation of eight databases housing genomic data from all four types of infectious pathogens—bacteria, viruses, eukaryotic pathogens like malaria, and disease-carrying vectors like mosquitoes.
The merging of bacterial and viral databases into BV-BRC offers better organization for analyses of bacterial hosts and infecting virus co-evolution. “These resources allow you to take your new bug and compare it to everything that’s currently in the database, and you can make some sense out of it. How similar is it? Are there any new genes in there, new variants of the genes in there? Is it something that is likely to be resistant or susceptible to antibiotic[s] or any other drug?” Stevens says.
Since the first human genome was sequenced in 2003, the ever-cheapening cost of sequencing has allowed researchers to harness data from labs across the globe and search across the genome in a few seconds. The combination of multiple sequencing efforts per species and collaborative efforts across the globe has amassed 200,000 bacterial genomes and 1.5 million viral genomes.
With the conglomeration of large amounts of data, scientists have now harnessed the power of machine learning in bacterial genomics. An algorithm can combine bacterial antibiotic resistance data with short DNA sequence data and build a correlative model of those DNA sequences. This creates a model that strongly predicts gene association with antibiotic resistance. Then, without additional information from biologists or annotations in the genomic data sets, the algorithm can take a new bacterial genome and predict its antibiotic susceptibility with up to 95 percent accuracy.
The algorithm’s findings rival traditional wet lab methods and are aiding efforts to develop new antibiotic therapeutics. As bacteria can evolve rapidly to gain antibiotic resistance while in a single human host, this algorithm has immediate utility in the clinic, as well as in biological research on bacterial evolution.
As for the evolution of machine learning and data science in biology, Stevens envisions its impact down many avenues in the near future. One potential clinical application is tabletop DNA sequencing platforms, which allow sequencing to run off a laptop. Stevens envisions that a clinician could input a patient’s bacterial swab, sequence their DNA, and enter the data into a predictive model. From there, the algorithm could output antibiotic resistance predictions based on a combination of mechanistic knowledge and machine learning from genomic resources such as BV-BRC.
In biology research, Stevens is working on automating complex analytical workflows. In this approach, the algorithm would present scientists with the results from a number of analytical processes, freeing them to work from a more “meta” level, pursuing whichever avenue presents the most interesting results. Stevens says that biology involves so much data, yet there are so many unknown mechanisms, making it particularly suited to data-driven models relative to physics or chemistry.
Stevens emphasizes that data processing is an iterative process that requires follow-up wet lab experiments. Big data could be integrated by, for example, feeding multifaceted clinical data into the computer algorithms, which find unexpected links between the number of early childhood virus infections and autism spectrum disorder diagnosis. Then, biologists form hypotheses about why immunosuppression and autism may be linked, and these mechanistic hypotheses are then tested at the lab bench.
The machine learning approach has highlighted previously ignored DNA sequences, such as non-coding DNA in mycobacterium tuberculosis which confers antibiotic resistance. In medicine, machine learning models are immediately useful, as they have been used to build predictive models for cancer treatment.
The conglomeration of large biological datasets has other benefits, such as allowing those outside the field, experts in computer science but not wet lab experiments, to partake in and contribute to biological research.
Stevens reflects on the opportunities for collaboration opened up by big data: “Biologists have always collaborated with each other, but with large-scale data sets, they now need new expertise in their lab. So, they need somebody who can do the computing.”
Stevens has witnessed that computer scientists can do more than offer their technical skills. “The really interesting collaborations are when the computer scientists are willing to ask questions that might be too stupid for the biologists to ask,” Stevens said. “Sometimes [those questions] turn out to be useful. I think what’s really important in these collaborations is to be really open to any method or idea or approach that can actually make some headway on a question…and it’s a really great fit for the way [U]Chicago works.”