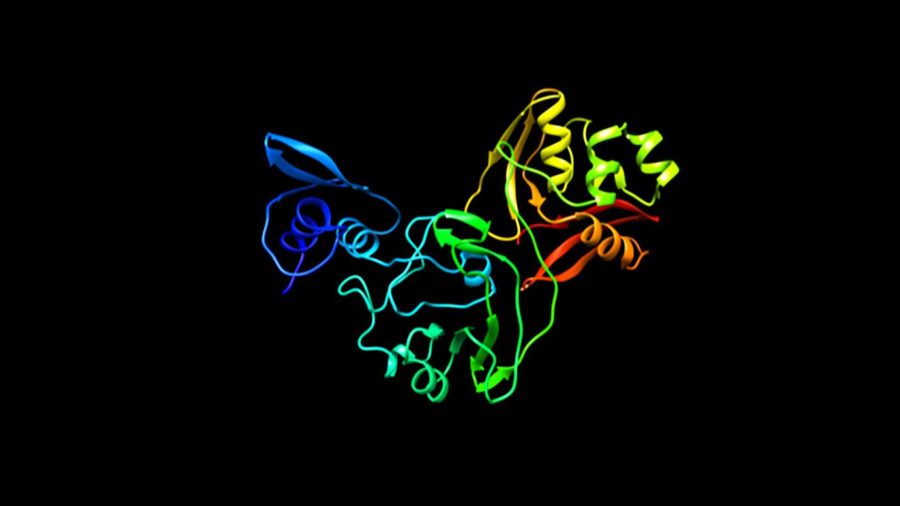
A newly mapped protein was identified in early March as a potential coronavirus drug target by a team of researchers from the University of Chicago, Argonne National Laboratory, Northwestern University Feinberg School of Medicine and the University of California, Riverside, School of Medicine.
At Argonne National Laboratory in Lemont, Illinois, researchers uncovered some protein structures from the SARS-CoV-2 virus. This virus is responsible for the coronavirus disease (COVID-19) outbreak that originated in Wuhan, China, and has developed into a global pandemic.
Proteins from SARS-CoV-2 were expressed, purified and crystallized at the Center for Structural Genomics of Infectious Diseases (CSGID), a research consortium that rapidly responds to emerging and unexpected infectious disease outbreaks.
Scientists have identified a protein in the virus similar to one in SARS-CoV, the virus that caused the 2002–03 SARS outbreak, and two proteases (which keep other proteins in the virus functioning correctly) that could be starting points or potential targets for a COVID-19 vaccine.
In late January, the National Institutes of Health (NIH) sent a sequence of the genome of the SARS-CoV-2 coronavirus to the CSGID research team at Northwestern University. There are 28 proteins in the virus’s genome, 16 of which have been identified as nonstructural proteins (Nsp) that form a complex responsible for regulating the virus’s replication.
“We focus on these nonstructural proteins because they form a large assembly that is involved in the replication of the virus.” Andrzej Joachimiak, co-director at the CSGID and a professor at the University of Chicago, said in an interview with The Maroon.
In early March, scientists mapped one protein, Nsp15, and found that it shares 89 percent similarity in terms of structure with a protein identified in SARS-CoV—the virus that caused an outbreak of SARS in 2002.
“If there were drugs for SARS Nsp15, we would have some drugs for this new virus now,” Joachimiak explained. “But unfortunately, we don’t have any vaccine or proven drugs.”
However, because of the similarity between SARS-CoV and SARS-CoV-2, the several inhibitors that were identified as effective against the SARS-CoV virus would provide “a good starting point for development of a potential inhibitor and eventually drug against SARS CoV-2”, Joachimiak and fellow Argonne scientist and researcher Robert Fischetti told The Maroon in a follow-up email. Fischetti serves as a group leader for Argonne’s X-ray Science Division and as a life sciences advisor to the director of Argonne’s Advanced Photon Source research facility.
However, they wrote, “it may take a year or two to develop an inhibitor into a drug that is effective and safe”.
In the sequenced genome, scientists also identified two proteases, enzymes which specifically cleave proteins. In coronaviruses, these proteases break down polyproteins to keep individual proteins functioning properly. However, it is possible to target these proteases’ ability to break down virus polyproteins and develop potential antivirals.
After determining the structures of the protein, scientists deposit them into the Protein Data Bank, allowing the entire scientific community to view and analyze the structures. CSGID researchers based at other universities are working to identify the structure of other proteins in SARS-CoV-2, including Nsp9, 10 and 16. On March 19, Northwestern University announced that researchers there had mapped Nsp10 and Nsp16.
After identifying the proteins’ structure, scientists can then begin working to identify inhibitors which can be used to block the production of proteins the virus needs to replicate. “If we can find inhibitors, we would determine the structure of the protein-inhibitor complexes, and try to use computational methods to design better inhibitors.” Joachimiak said.
This method, structure-based drug discovery, could allow scientists to develop a drug based on the structure of the protein. Through the screening of chemical libraries, scientists can identify potential inhibitors that bind to the protein and are targets for three-dimensional structure determination. Comparing those to the structure of the complex, the team can effectively improve the binding and develop inhibitors.
Joachimiak also acknowledged possibly collaborating with pharmaceutical companies in the future. “They can help us with biochemical assays for binding,” he said. “We would like to understand the interactions with small molecules, and those companies may have some libraries of the unique compounds for discovering novel inhibitors.”
CSGID was established in 2007. It is funded by the NIH and the National Institute of Allergy and Infectious Diseases. X-ray diffraction data was collected at the Advanced Photon Source, funded by the U.S. Department of Energy.
Since its establishment, scientists at the CSGID have determined the three-dimensional structure of over 1,200 proteins from bacteria and viruses, a step which is critical in the development of counteractive drugs and vaccines.